Driven to the past by a frightening future
I am thankful that I have witnessed some of the most significant advances in technology in my lifetime; moon landing, the internet, smartphone and DNA analysis just to name a few.
Nucleosomes are packed together to form chromatin fibers and chromosomes.
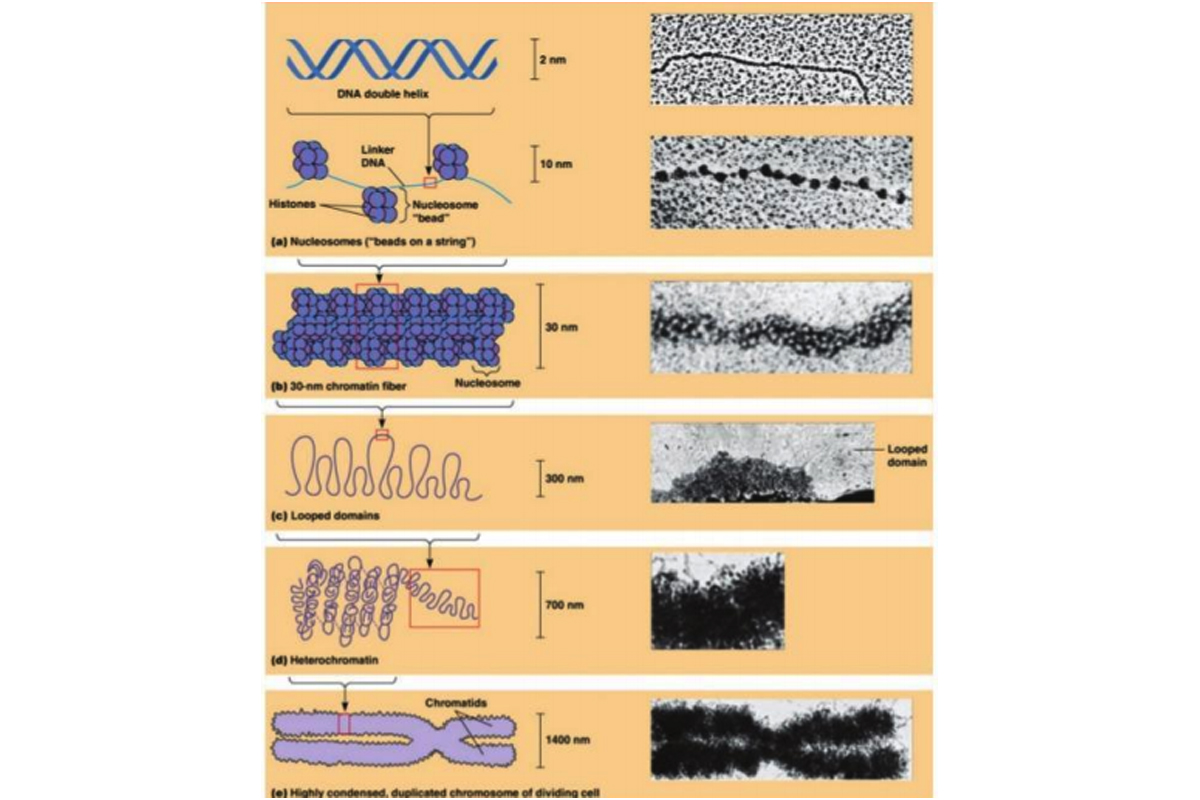
The formation of nucleosomes is only the first step in the packaging of nuclear DNA. Isolated chromatin fibers exhibiting the “beadson-a-string” appearance measure about 10 nanometre in diameter, but the chromatin of intact cells often forms a slightly thicker fiber about 30 nm in thickness called the 30-nm chromatin fiber.
In preparations of isolated chromatin, the 10-nm and 30-nm forms of the chromatin fiber can be interconverted by changing the salt concentration of the solution.
However, the 30-nm fiber does not form in chromatin preparations whose histone H1 molecules have been removed, suggesting that histone H1 facilitates the packing of nucleosomes into the 30-nm fiber.
Advertisement
Several models have been proposed to explain how individual nucleosomes are packed together to form a 30-nm fiber.
Most early models postulated that the chain of nucleosomes is twisted upon itself to form some type of coiled structure.
However, recent studies suggest that the struc-ture of the 30- nm fiber is much less uniform than a coiled model suggests. Instead, the nucleosomes of the 30-nm fiber seem to be packed together to form an irregular, three-dimensional zigzag structure that can interdigitate with its neighbouring fibers.
The next level of chromatin packaging is the folding of the 30-nm fibers into looped domains averaging 50,000-100,000 bp in length.
This looped arrangement is maintained by the periodic attachment of DNA to an insoluble network of nonhistone proteins, which form a chromosomal scaffold to which the long loops of DNA are attached.
The looped domains can be most clearly seen in electron micrographs of chromosomes isolated from dividing cells and treated to remove all the histones and most of the nonhistone proteins.
Loops can also be seen in specialised types of chromo-somes that are not associated with the process of cell division. In these cases, the chromatin loops turn out to contain “active” regions of DNA – that is, DNA that is being transcribed. It makes sense that active DNA would be less tightly packed than inactive DNA because it would allow easier access by proteins involved in gene transcription. Even in cells, where genes are being actively tran-scribed, significant amounts of chromatin may be further compacted.
The degree of folding in such cells varies over a continuum. Segments of chromatin so highly compacted that they show up as dark spots in micrographs are called heterochromatin, whereas the more loosely packed, diffuse form of chromatin is called euchromatin.
The tightly packed heterochromatin contains DNA that is transcriptionally inactive, while the more loosely packed euchromatin is associated with DNA that is being actively transcribed.
Much of the chromatin in metabolically active cells is loosely packed as euchromatin, but as a cell prepares to divide, all of its chromatin becomes highly compacted, generating a group of microscopically distinguishable chromosomes. Because the chromosomal DNA has been recently duplicated, each chromosome is composed of two duplicate units called chromatids.
The extent to which a DNA molecule has been folded in chromatin and chromosomes can be quantified using the DNA packing ratio, which is calculated by determin-ing the total extended length of a DNA molecule and dividing it by the length of the chromatin fiber or chromo-some into which it has been packaged.
The initial coiling of the DNA around the histone cores of the nucleosomes re-duces the length by a factor of about seven, and formation of the 30-nm fiber results in a further sixfold condensa-tion.
The packing ratio of the 30-nm fiber is therefore about 7×6 = 42.
Further folding and coiling brings the overall packing ratio of typical euchromatin to about 750.
For heterochromatin and the chromosomes of dividing cells, the packing ratio is still higher. At the time of cell division, for example, a typical human chromosome mea-sures about 4-5 mm in length, yet contains a DNA mole-cule that would measure almost 75 mm if completely extended.
The packing ratio for such a chromosome there-fore falls in the range of 15,000-20,000.
The writer is associate professor and head, department of botany, Ananda Mohan College, Kolkata.
Advertisement