Maharashtra bus accident: Bodies to be identified through DNA testing
According to officials, the bus was travelling from Maharashtra's Yavatmal to Pune and met with an accident in Buldhana on the Samruddhi Mahamarg Expressway.
Nucleosomes are the basic units of a chromatin structure.
Representational Image.
The DNA contained within a typical nucleus would measure a metre or more in length if it were completely extended, whereas the nucleus itself is usually no more than five to 10 µm in diameter. The folding of such an enormous length of DNA into a nucleus that is almost a million times smaller presents a significant topological problem.
One of the first insights into the folding process emerged in the late 1960s, when X-ray diffraction studies carried out by Maurice Wilkins revealed that purified chromatin fibres have a repeating structural subunit that is seen in neither DNA nor histones alone. Wilkins therefore concluded that histones impose a repeating structural organisation upon DNA.
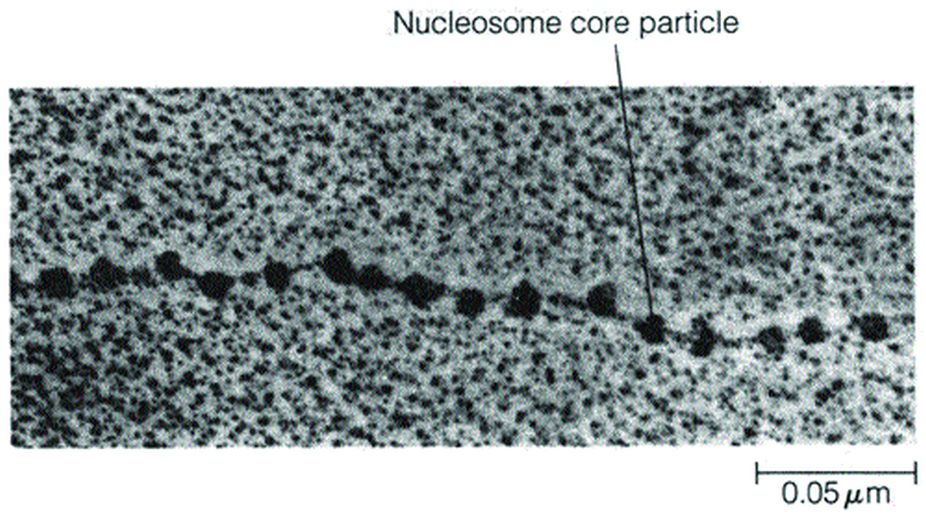
A clue to the nature of this structure was provided in 1974, when Ada Olins and Donald Olins published electron micrographs of chromatin fibres isolated from cells in a way that avoided the harsh solvents used in earlier procedures for preparing chromatin for microscopic examination. Chromatin fibres viewed in this way appear as a series of tiny particles attached to one another by thin filaments.
Advertisement
This “beads-on-a-string” appearance led to the suggestion that the beads are made of proteins (presumably histones) and the thin filaments connecting the beads correspond to DNA. We now refer to each bead, along with its associated short stretch of DNA, as a nucleosome.
On the basis of electron microscopy alone, it would have been difficult to determine whether nucleosomes are a normal component of chromatin or an artifact generated during sample preparation. Fortunately, independent evidence for the existence of a repeating structure in chromatin was reported at about the same time by Dean Hewish and Leigh Burgoyne, who discovered that rat liver nuclei contain a nuclease that is capable of cleaving the DNA in chromatin fibres.
In one crucial set of experiments, these investigators exposed chromatin to this nuclease and then purified the partially degraded DNA to remove chromatin proteins. When they examined the purified DNA by gel electrophoresis, they found a distinctive pattern of fragments in which the smallest piece of DNA measured about 200 bp in length, and the remaining fragments were exact multiples of 200 bp.
Since nuclease digestion of protein-free DNA does not generate this fragment pattern, they concluded that first, chromatin proteins are clustered along the DNA molecule in a regular pattern that repeats at intervals of roughly 200 bp, and second, the DNA located between these protein clusters is susceptible to nuclease digestion, yielding fragments that are multiples of 200 bp in length.
The question arose whether the protein clusters postulated to occur at 200 bp intervals correspond to the spherical particles observed in electron micrographs of chromatin fibres. Answering this question required a combination of the nuclease digestion and electron microscopic approaches.
Chromatin was briefly exposed to micrococcal nuclease, a bacterial enzyme that, like the rat liver nuclease, cleaves chromatin DNA at intervals of 200 bp.
The fragmented chromatin was then separated into fractions of varying sizes by centrifugation and examined by electron microscopy. The smallest fraction was found to contain single spherical particles, the next fraction contained clusters of two particles, the succeeding fraction contained clusters of three particles, and so forth.
The writer is associate professor, head, department Of botany, ananda mohan college, kolkata, and also Fellow, botanical society of bengal, and can be Contacted at tapanmaitra59@yahoo.co.in
Advertisement