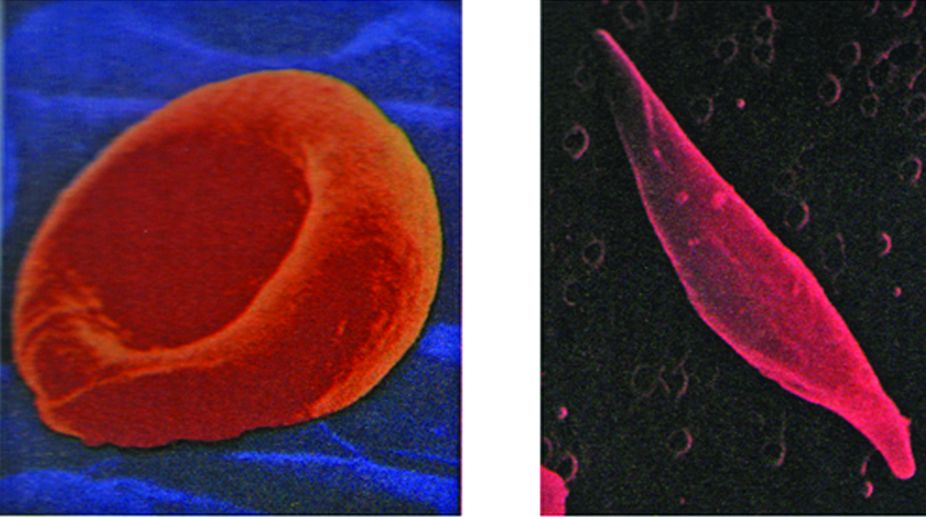
The theory that genes direct the production of enzyme molecules represented a major advance in our understanding of gene action, but it provided little insight into the question of how genes accomplish this task. The first clue to the underlying mechanism emerged a few years later in the laboratory of Linus Pauling, who was studying the inherited disease sickle-cell anaemia. The red blood cells of individuals suffering from sickle-cell anaemia exhibit an abnormal, “sickle” shape that causes the cells to become trapped and damaged when they pass through small blood vessels.
In trying to identify the reason for this behaviour, Pauling decided to analyse the properties of haemoglobin, the major protein of red blood cells. Because haemoglobin is a charged molecule, he used the technique of electrophoresis, which separates charged molecules from one another by placing them in an electric field. Pauling found that haemoglobin from sickle cells migrated at a different rate than normal haemoglobin, suggesting that the two proteins differ in electric charge. Since some amino acids have charged side chains, Pauling proposed that the difference between normal and sickle-cell haemoglobin lay in their amino acid compositions.
To test this hypothesis, it seemed necessary to know the entire amino acid sequence of haemoglobin. At the time of Pauling’s discovery in the early 1950s, the largest protein to have been sequenced was less than one-tenth the size of haemoglobin, so determining the complete amino acid sequence of haemoglobin would have been a monumental undertaking.
Fortunately, an ingenious shortcut devised by Vernon Ingram made it possible to identify the amino acid abnormality in sickle-cell haemoglobin without determining the protein’s complete amino acid sequence. Ingram used the protease trypsin to cleave haemoglobin into peptide fragments, which were then separated from each other. When Ingram examined the peptide patterns of normal and sickle-cell haemoglobin, he discovered that only one peptide differed between the two proteins.
Analysis of the altered peptide revealed that a glutamic acid in normal haemoglobin had been replaced by a valine in sickle-cell haemoglobin. Since glutamic acid is negatively charged and valine is neutral, this substitution explains the difference in electrophoretic behaviour between normal and sickle-cell haemoglobin originally observed by Pauling. The discoveries by Pauling and Ingram necessitated several refinements in the one gene-one enzyme concept of Beadle and Tatum.
First, the fact that haemoglobin is not an enzyme indicates that genes encode the amino acid sequences of proteins in general, not just enzymes. In addition, the discovery that different genes code for the a and j3 chains of haemoglobin reveals that each gene encodes the sequence of a polypeptide chain, not necessarily a complete protein. Thus, the original hypothesis was refined into the one gene-one polypeptide theory. According to this theory, the nucleotide sequence of a gene determines the sequence of amino acids in a polypeptide chain.
In the mid-1960s, this prediction was confirmed in the laboratory of Charles Yanofsky, where the locations of dozens of mutations in the bacterial gene coding for a subunit of the enzyme tryptophan synthase were determined. As predicted, the positions of the mutations within the gene correlated with the positions of the resulting amino acid substitutions in the tryptophan synthase polypeptide chain. Showing that a gene’s base sequence specifies the amino acid sequence of a polypeptide chain represented a major milestone, but subsequent developments have revealed that gene function is often more complicated than this, especially in eukaryotes.
Most eukaryotic genes contain non-coding sequences interspersed among the coding regions and therefore do not exhibit a complete linear correspondence with their polypeptide product. Moreover, the coding sequences contained within such genes can be read in various combinations to produce different mRNAs, each coding for a unique polypeptide chain. This phenomenon, called alternative RNA splicing, allows dozens or even hundreds of different polypeptides to be produced from a single gene.
There are several types of genes that do not produce polypeptide chains at all. These genes code for RNA molecules such as transfer RNAs, ribosomal RNAs, small nuclear RNAs, and microRNAs, each of which performs a unique cellular function.
Thus the one gene-one polypeptide view of gene function has become obsolete, replaced by a broader view in which genes are seen as functional units of DNA that code for the amino acid sequence in one or more polypeptide chains, or alternatively, for one of several types of RNA that perform functions other than specifying the amino acid sequence of polypeptide chains.
(The writer is associate professor, head, department of botany, ananda mohan college, kolkata, and also fellow, botanical society of bengal, and can be contacted at tapanmaitra59@yahoo.co.in)